Gene Regulatory Networks - Logic Networks¶
Description¶
Gene regulatory networks are useful models for biologists to understand the interactions among genes in living organisms. In this work, we consider a special class of gene regulatory networks, called logical networks, where the “links” among genes are inferred by logic analysis of phylogenetic profiles. We provide a formalization of logic networks, and introduce a membrane computing model for reconstructing logical networks and reproducing their dynamics. An example logical network is simulated by using software P-Lingua and MeCoSim. The simulation result shows that membrane computing is a versatile framework for modeling different biological scenarios.
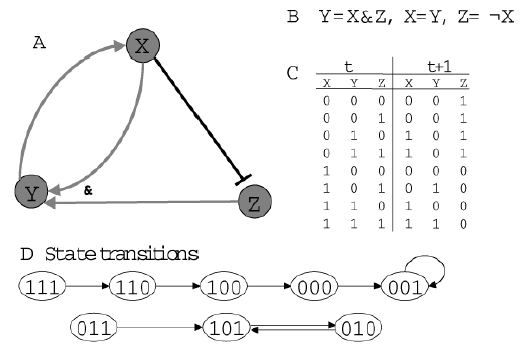
Model¶
The corresponding P-Lingua file is the following:
@model<probabilistic>
def main()
{
@mu = []'global;
@mu(global) += [ []'1 ]'{100+k},{100+k} : 1<=k <= (ng+nb+nu);
{
@ms({g{k,1}+100}) += a{1}*g{k,3},a{0}*(1-g{k,3});
@ms({g{k,1}+100}) += go;
} : 1<=k<=ng;
{
@ms({t{i,1}+100+ng}) += binop{t{i,2}};
} : 1<=i<=nb;
{
@ms({unt{i,1}+100+ng+nb}) += unop{unt{i,2}}; /* unt: unary table; unop: unary operator; nu: number of unary operators*/
} : 1<=i<=nu;
/* Self-influence of a gen over itself. */
/*rs{1,i}*/ go,a{i}[]'1 --> c{i},b{i}*(max*i),b{0}*threshold,clock{0}[]'1 :: 1 : 0<=i<=1;
/* For each gen, an object is created */ /* Unary operators are included. */
/*re{2,i,j,k}*/
{
[c{i}]'{100+g{j,1}} --> [&{auxDest{i,g{j,1},l}}:{1<=l<=nb+nu}]'{100+g{j,1}} :: 1;
} : 1<=j<=ng, 0<=i<=1;
/*re{3,i,j,k}*/
{
[auxDest{i,g{j,1},k}]'{100+g{j,1}} --> [dest{i,g{j,1},t{k,1}+ng}]'{100+g{j,1}} :: 1;
} : 1<=j<=ng, 0<=i<=1, 1<=k<=nb;
/*re{4,i,j,k}*/
/* Spread the effect to unary operators (promoters and inhibitors). */
{
[auxDest{i,g{j,1},k}]'{100+g{j,1}} --> [dest{i,g{j,1},unt{k-nb,1}+ng+nb}]'{100+g{j,1}} :: 1;
} : 1<=j<=ng, 0<=i<=1, nb+1<=k<=nb+nu;
/* Creation of objects representing real interactions. */
{
/*re{5,i,k}*/ [dest{i,t{k,3},t{k,1}+ng} --> e{(t{k,4}*i+(1-i)*(1-t{k,4})),t{k,1}+ng}]'{100+t{k,3}} :: 1;
/*re{6,i,k}*/ [dest{i,t{k,5},t{k,1}+ng} --> e{(t{k,6}*i+(1-i)*(1-t{k,6})),t{k,1}+ng}]'{100+t{k,5}} :: 1;
} : 1<=k<=nb, 0<=i<=1;
/* For unary operators. */
/*re{7,i,k}*/
{
[dest{i,unt{k-nb,3},unt{k-nb,1}+ng+nb} --> e{(unt{k-nb,4}*i+(1-i)*(1-unt{k-nb,4})),unt{k-nb,1}+ng+nb}]'{100+unt{k-nb,3}} :: 1;
} : nb+1<=k<=nb+nu, 0<=i<=1;
/* Send gene data to interactions environments. */
{
/*re{8,i,k}*/ [[]'{100+t{k,1}+ng}[e{i,t{k,1}+ng}]'{100+t{k,3}} --> [a{i}]'{100+t{k,1}+ng}[]'{100+t{k,3}}]'global :: 1;
/*re{9,i,k}*/ [[]'{100+t{k,1}+ng}[e{i,t{k,1}+ng}]'{100+t{k,5}} --> [a{i}]'{100+t{k,1}+ng}[]'{100+t{k,5}}]'global :: 1;
} : 1<=k<=nb, 0<=i<=1;
/* For unary operators. */
/*re{10,i,k}*/
{
[[]'{100+unt{k-nb,1}+ng+nb}[e{i,unt{k-nb,1}+ng+nb}]'{100+unt{k-nb,3}} --> [a{i}]'{100+unt{k-nb,1}+ng+nb}[]'{100+unt{k-nb,3}}]'global :: 1;
} : nb+1<=k<=nb+nu, 0<=i<=1;
/*OR interactions*/
/*rs11*/ a{0}*2,binop{1} []'1 --> binop{1},c{0}[]'1 :: 1;
/*rs12*/ a{1}*2,binop{1} []'1 --> binop{1},c{1}[]'1 :: 1;
/*rs13*/ a{1},a{0},binop{1} []'1 --> binop{1},c{1}[]'1 :: 1;
/*AND interactions*/
/*rs14*/ binop{2},a{1}*2[]'1 --> binop{2},c{1}[]'1 :: 1;
/*rs15*/ binop{2},a{0}*2[]'1 --> binop{2},c{0}[]'1 :: 1;
/*rs16*/ binop{2},a{1},a{0}[]'1 --> binop{2},c{0}[]'1 :: 1;
/*XOR*/
/*rs17*/ binop{3},a{1}*2[]'1 --> binop{3},c{0}[]'1 :: 1;
/*rs18*/ binop{3},a{0}*2[]'1 --> binop{3},c{0}[]'1 :: 1;
/*rs19*/ binop{3},a{1},a{0}[]'1 --> binop{3},c{1}[]'1 :: 1;
/*NXOR*/
/* These rules are finally removed, having being considered unnecesary given the possibility of negating the output of any of the previous interaction types (in particular, XOR). */
/*rs20*/ /*binop{4},a{1}*2[]'1 --> binop{4},c{1}[]'1 :: 1;*/
/*rs21*/ /*binop{4},a{0}*2[]'1 --> binop{4},c{1}[]'1 :: 1;*/
/*rs22*/ /*binop{4},a{1},a{0}[]'1 --> binop{4},c{0}[]'1 :: 1;*/
/* For promoters. Remember: (unop: unary operator) (binop: ternary operator)*/
{
/*rs23*/ unop{1},a{i}[]'1 --> unop{1},c{i}[]'1 :: 1;
/*rs24*/ unop{2},a{i}[]'1 --> unop{2},c{1-i}[]'1 :: 1;
/*rs25*/ unop{3},a{i} []'1 --> unop{3},c{i}*i[]'1 :: 1;
/*rs26*/ unop{4},a{i} []'1 --> unop{4},c{1-i}*i[]'1 :: 1;
} : 0<=i<=1;
/* Operation inside the operator: */
{
/*re27*/[[c{i}]'{100+t{k,1}+ng}[]'{100+t{k,7}} --> []'{100+t{k,1}+ng}[eF{(t{k,8}*i+(1-i)*(1-t{k,8})),(t{k,1}+ng)}]'{100+t{k,7}}]'global :: 1;
/*rs29*/eF{i,(t{k,1}+ng)}[]'1 --> b{i}*t{k,9}[]'1 :: 1;
} : 0 <= i <=1, 1<=k<=nb;
/* Promoters and inhibitors: */
{
/*re28*/[[c{i}]'{100+unt{k,1}+ng+nb}[]'{100+unt{k,5}} --> []'{100+unt{k,1}+ng+nb}[eF{i,(unt{k,1}+ng+nb)}]'{100+unt{k,5}}]'global :: 1;
/*rs30*/eF{i,(unt{k,1}+ng+nb)}[]'1 --> b{i}*unt{k,6}[]'1 :: 1;
} : 0 <= i <=1, 1<=k<=nu;
/* Rules to remove b{i} objects. */
/* rs31 */
b{1},b{0}[]'1 --> #[]'1 :: 1;
/*Clock control: */
/*rs32{i}*/
clock{i-1}[]'1 --> clock{i}[]'1 :: 1 : 1<=i<=cc;
/* In this moment all contributions to each gene have been received, neutralizing complementary pairs of influences, so we force a change in the charge to activate the rules evaluating the state when the current cycle is over. */
/*rs33*/
clock{cc}[]'1 --> clock{cc+1}-[]'1 :: 1;
/* Objects b{0} go into d{0}, thus making working alphabet objects be different from the ones in the environment.*/
/*rs34*/
b{0}-[]'1 --> -[d{0}]'1 :: 1;
/*rs35*/
b{1}-[]'1 --> -[]'1 :: 1;
/* Clock progress until cc+2*/
/*re36{i,j}*/
[clock{cc+i} --> clock{cc+i+1}]'{100+g{j,1}} :: 1 : 1<=j<=ng, 1<=i<=2;
/* This rule is removed because it finally has no sense, given that clock objects are only created in the environments containing go, that is, in the environments representing genes.
[clock{cc+j} --> clock{cc+j+1}]'{100+t{k,1}+ng} :: 1 : 1<=k<=nb, 1<=j<=2;*/
/* Remaining dest objects removal. */
/*rs37{i,j,k}*/
{
dest{i,j,t{k,1}+ng}-[]'1 --> -[]'1 :: 1;
} : 1<=j<=ng, 0<=i<=1, 1<=k<=nb;
/* For promoters and inhibitors: */
/*rs38{i,j,k}*/
{
dest{i,j,unt{k-nb,1}+ng+nb}-[]'1 --> -[]'1 :: 1;
} : 1<=j<=ng, 0<=i<=1, nb+1<=k<=nb+nu;
/* If there were remaining objects implying negative contribution,
the charge of the membrane is changed in order to distinguish the possible states. */
/*rs39*/
-[d{0}]'1 --> +[]'1 :: 1;
/* Depending of the charge of membrane labelled by 0, the state of the gene is set to enabled (1) or disabled (0). In addition, the corresponding go objects are generated, in order to enable the start of the next cycle.*/
clock{cc+3}+[]'1 --> go,a{0}[]'1 :: 1;
clock{cc+3}-[]'1 --> go,a{1}[]'1 :: 1;
}
Files¶
- Custom application.
- Model file.
- Scenario files: simple network example, arabidopsis thaliana.
Results¶
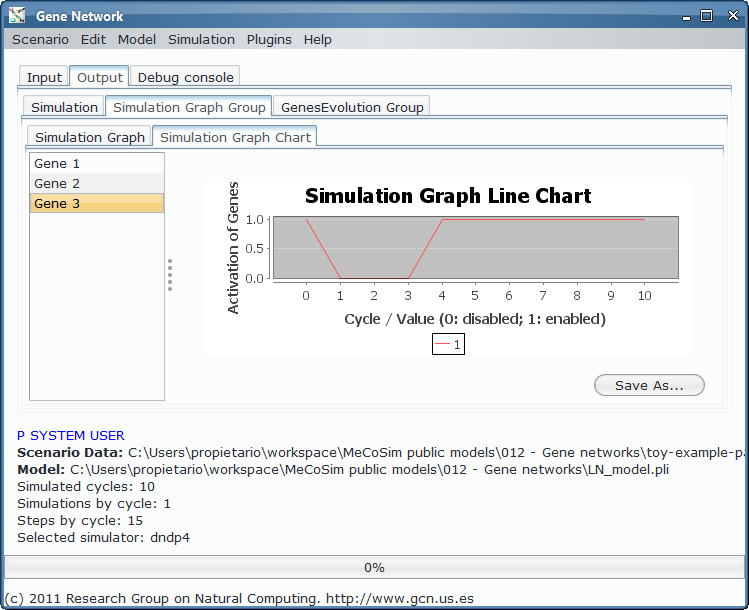
The evolution takes the data from a specific network introduced by the user and gives as the evolution of the network through the desired cycles. The toy example network evolves in some few steps as shown in the picture:
Arabidopsis case study¶
In order to experimentally verify our model, we have tested our algorithm by using a relatively large logic network. The data from this network has been obtained from NCBI/EBI database. The logic network has been constructed according to the procedure described by Bowers. This network regulates owering processes associated to the species Arabidopsis thaliana on a long day scenario. A. thaliana is a species widely used in genetic and protein interaction networks. The total number of genes in the network is 29, whereas the total number of interactions is 99. These interactions consist of 23 unary interactions and 76 binary interactions.
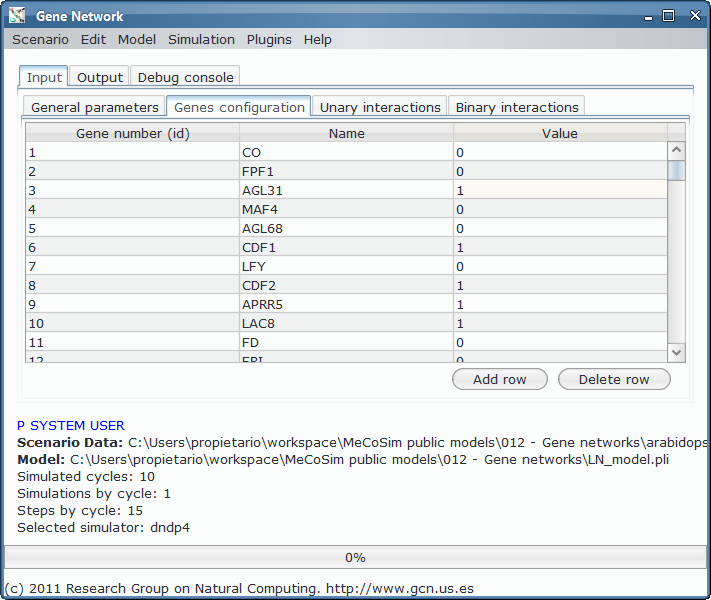
The proper initial states and data about unary and binary interactions are given:
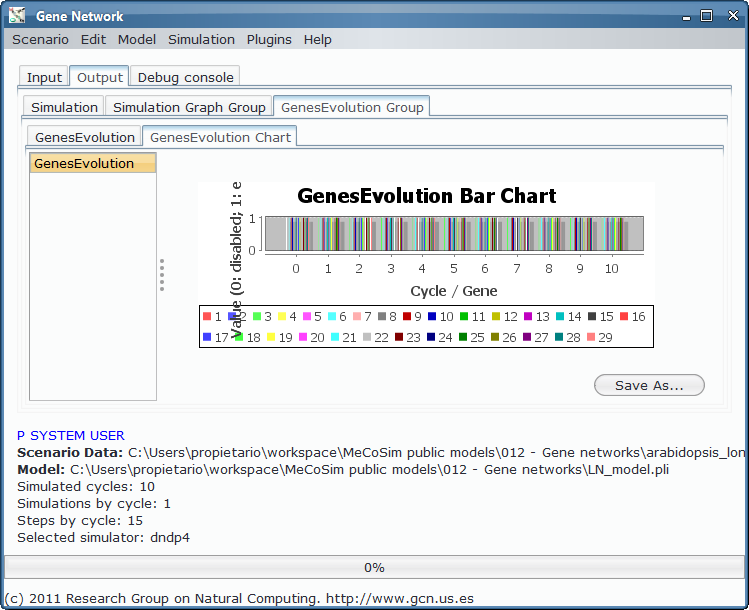
Finally, the evolution is shown in different output tables, line and bar charts:
References¶
- Analysing Gene Networks with PDP Systems. Arabidopsis thaliana, a Case Study (detailed here).
- Modeling Logic Gene Networks by Means of Probabilistic Dynamic P Systems, International Journal of Unconventional Computing (detailed here).
- Bowers, P.M., Cokus, S.J., Yeates, T.O., Eisenberg, D.: Use of logic relationships to decipher protein network organization. Science 5705(306), 2246-2249 (2004).